Genetic & demographic monitoring
What is genetic monitoring?
Adapted from Wikipedia, genetic monitoring is the use of molecular genetic markers to (i) identify and monitor individuals, species or populations, or (ii) to quantify changes in population genetic metrics (such as effective population size, genetic diversity, and population size) over time, (iii) detect change in frequency of adaptive molecular variation (Fig. 1). Genetic monitoring can thus be used to detect changes in species abundance and/or diversity, and has become an important tool in conservation.
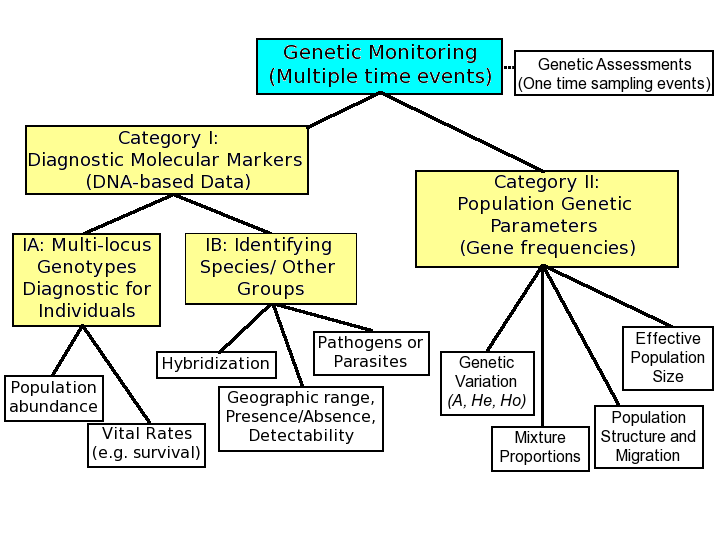
(Above) Categories of genetic monitoring. Category I includes the use of diagnostic molecular markers for traditional population monitoring through the identifi cation of individuals and species, and the repeated (temporal) assessment of population size. Category II includes the use of genetic markers to monitor population genetic parameters. Category III involves use of DNA markers to detect changes in frequency of adaptive alleles or gene expression associated with environmental change. From Allendorf et al. (2013), Modified from Schwartz et al. (2007).
Why is genetic monitoring important?
It is crucial to estimate and monitor the effective number of breeders (Nb) in natural populations to allow early detection of population declines. Early detection of a decline can help prevent loss of genetic diversity, population extirpation, and loss of ecosystem services often provided by local populations. Nb monitoring also allows detection of population growth or expansion following species restoration or spread of invasive species (Tallmon et al. 2012).
